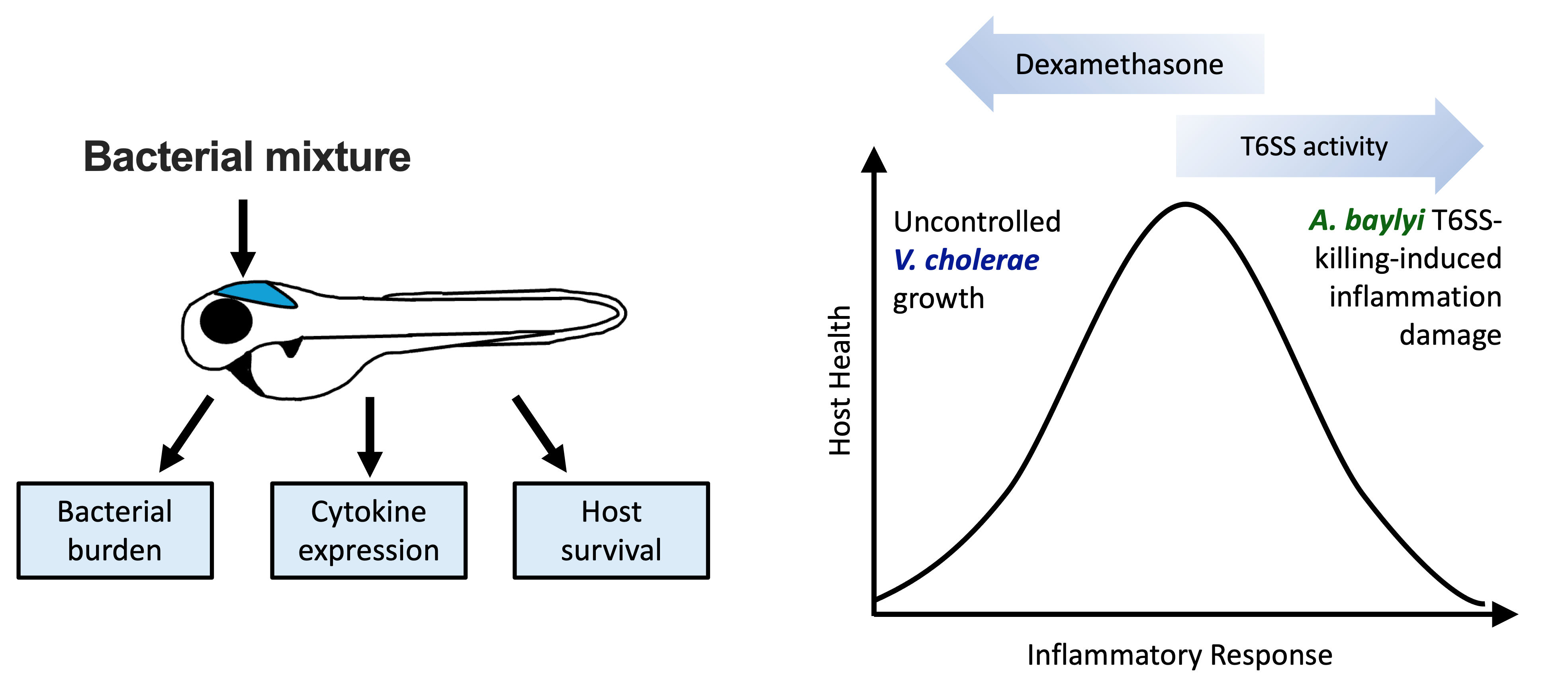
 For the past 15 years at UCL, the Thalassinos Lab has been studying alpha-1-antitrypsin, focusing on understanding the early misfolding events and how monomers form higher-order oligomers. Despite numerous models reported in the literature, the formation of the dimer as the first step towards higher-order oligomers has always been intriguing, with many different models of the dimer being reported.
For the past 15 years at UCL, the Thalassinos Lab has been studying alpha-1-antitrypsin, focusing on understanding the early misfolding events and how monomers form higher-order oligomers. Despite numerous models reported in the literature, the formation of the dimer as the first step towards higher-order oligomers has always been intriguing, with many different models of the dimer being reported.
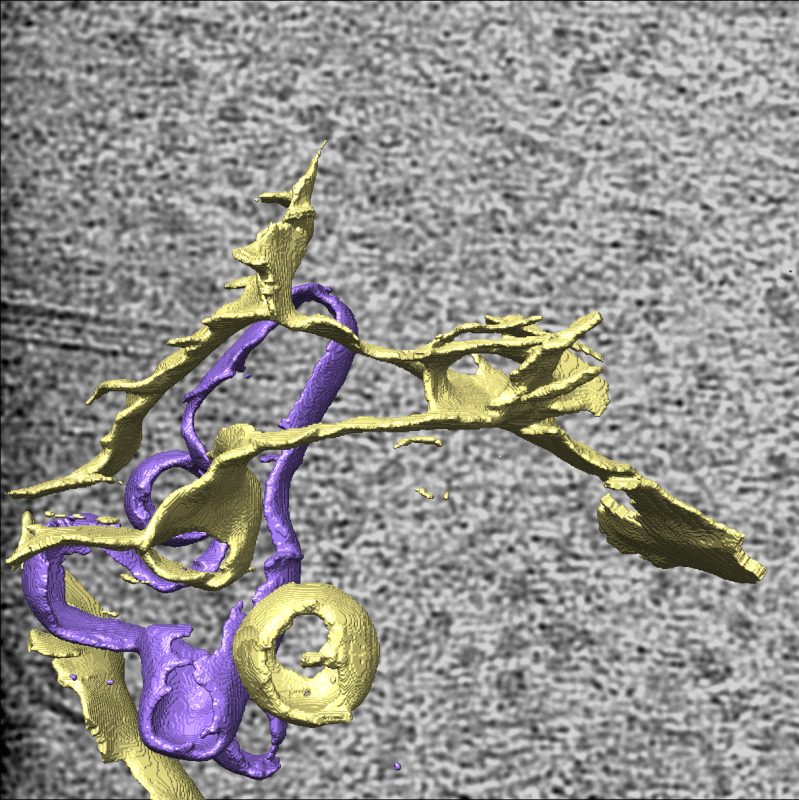
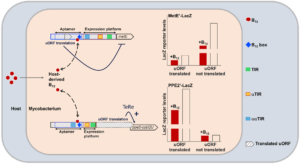
Using a combination of cyclic ion mobility and top-down electron capture dissociation, the team has identified and characterised an intermediate misfolded metastable state, where approximately 12% of the molecule at the C-terminus is displaced. Their data supports a polymer mediated by an intermolecular C-terminal domain insertion but also establishes a previously unobserved progression of pathogenic structural changes, thus extending the mechanism of alpha-1-antitrypsin polymerization. Importantly, this intermediate was also observed in alpha-1 protein extracted from human tissue.
Previous proposed intermediates were extrapolated from in vitro studies using methods that study the bulk average. The unique ability of ion mobility mass spectrometry to separate co-existing conformers and perform native top-down fragmentation, along with performing these analyses on ex vivo material, has been crucial. This highlights the power of ion mobility for studying early misfolding events.
The Thalassinos Lab extends their gratitude to the Irving and Lomas labs for their collaboration, and all the patients who provided material for this study.
For more information: https://pubs.acs.org/doi/epdf/10.1021/jacs.4c18139?ref=article_openPDF
 A new project led by UCL seeks to develop the plants and food of the future, supported by UK government funding from the Advanced Research and Invention Agency (ARIA).
A new project led by UCL seeks to develop the plants and food of the future, supported by UK government funding from the Advanced Research and Invention Agency (ARIA).