Structure/function studies of membrane proteins (primarily ion channels) and on bioinformatics and methods development for circular dichroism spectroscopy.
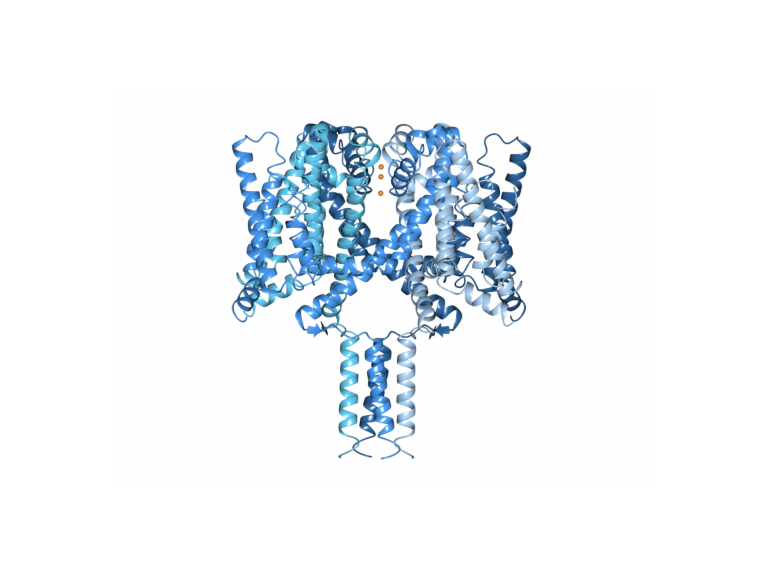
My lab focuses on structure/function studies of membrane proteins, especially those involved in ion translocation and transport. Of particular interest are voltage-gated sodium channels: we have determined the first crystal structure of a sodium channel in its open conformation and the first complete structure of any sodium channel. These have provided novel insights into the structural basis of channel gating and the molecular basis of diseases and drug binding, and have identified for the first time the locations of sodium ions in the channel. My lab uses the techniques of X-ray crystallography, cryo electron microscopy, circular dichroism spectroscopy, cloning and expression, bioinformatics, and molecular modelling, as well as functional studies to examine structure/function relationships.
The other main focus of my lab is the development of techniques and applications for circular dichroism (CD) spectroscopy, including synchrotron radiation circular dichroism (SRCD) spectroscopy of proteins, and special techniques required for circular dichroism spectroscopy of membrane proteins. We have created a number of widely-used bioinformatics tools, including the DichroWeb server for secondary structure analyses, the Protein Circular Dichroism Data Bank (PCDDB) repository for CD spectral and metadata, new soluble and membrane protein (and the soon to be available intrinsically-disordered protein) reference databases for CD analyses, the DichroMatch structural analysis server, and the CD data processing and archive package "CDTool".