Finn Werner’s group published a paper in Nature Communications on 22nd February.
The paper studies the mechanism of how viruses enter and replicate in the hosts cells is of fundamental importance to understand how they cause disease and developing tools for control.
A team of scientists at UCL, led by Prof Finn Werner, have taken a major step forward in understanding how African swine fever virus (ASFV) genes are controlled are expressed. ASFV causes a fatal disease of domestic pigs and wild boar that results in severe socio-economic impacts in affected countries in Africa Europe, Asia and parts of Oceana and the Caribbean. The lack of tools including vaccines or antivirals limits control of disease.
ASFV replicates in the host cell cytoplasm and uses its own machinery to transcribe its genes into mRNAs which are translated into proteins required for virus replication or modulating host cell function. The UCL team have expressed and assembled 8 proteins comprising the ASFV RNA polymerase into an active complex. The cryo-EM structure of this complex molecular machine was determined providing the information to further probe how it functions to regulate ASFV gene expression. Dr Pilotto says: ‘The production of recombinant eukaryotic RNA polymerases remains the bottleneck for structural studies and high-throughput inhibitor screenings. Our success with the ASFV RNAP represents a game-changer in the field’. The ASFV RNAP bears a striking resemblance to RNAPII – and key differences include the fusion of the ‘assembly platform subunits’ and an unusual fusion with a domain related to the eukaryotic mRNA capping enzyme. Together, the changes represent adaptions to streamline the enzyme to serve the virus best: allowing for efficient RNAP biogenesis, and mRNA expression and processing. The availability of a recombinant functional RNA polymerase will facilitate high throughput screening of antiviral compounds to identify compounds with sufficient specificity and selectivity.
This research will be further developed in a recently funded BBSRC collaborative research project between UCL and The Pirbright Institute (led by Drs Linda Dixon and Chris Netherton). This project will identify other accessory virus and host factors involved in regulating the temporal expression of ASFV genes and the packaging of the virus RNA polymerase into particles ready to start a next round of infection. This information is critical to understand the ASFV replication cycle and potential host factors that may limit virus replication.
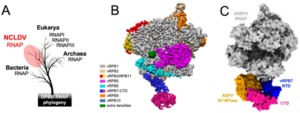
Evolution and structure of ASFV RNAP. All DPBB RNAP share a common ancestry (A), the cryoEM structure of the 8-subunit ASFV RNAP (B), and model of the RNAP-capping enzyme complex showing the interaction with the CE (N7-MTase in orange, vRPB7 in blue/magenta) 1. ASFV research in the RNAP lab is funded by the BBSRC (BB/X015424/1) and Wellcome Trust (108877/B/15/Z).
Reference
- Pilotto, S., Sykora, M., Cackett, G., Dulson, C., and Werner, F. (2024). Structure of the recombinant RNA polymerase from African Swine Fever Virus. Nat Commun 15, 1606. 10.1038/s41467-024-45842-7.
Full paper can be read here.