
Structural Immunology Group
Structural Immunology Group
Our major research interests lie in solution structural determinations of proteins and their interactions in the immune system, primarily involving the complement proteins and antibodies. Our major technologies include small angle X-ray and neutron solution scattering at Diamond and ISIS (SAXS, SANS) and analytical ultracentrifugation (AUC) on our Beckman Optima and two Beckman XL-I analytical ultracentrifuges, one of which is equipped with fluorescent optics. We also possess surface plasmon resonance and dual polarization interferometry instruments that help with our interactions studies, access protein crystallography through collaborations. We also maintain web sites that detail the variants and phenotypes in mutant complement and coagulation proteins (such as that for Haemophilia B - https://www.factorix.org/) (Xu et al., 2023). In the UK, we are involved with the CCP-SAS project, a joint US/UK collaborative open-source software development project that provides the infrastructure and tools for analysing small angle scattering data on complex systems using atomistic and coarse grained modelling approaches (see http://www.ccpsas.org/).
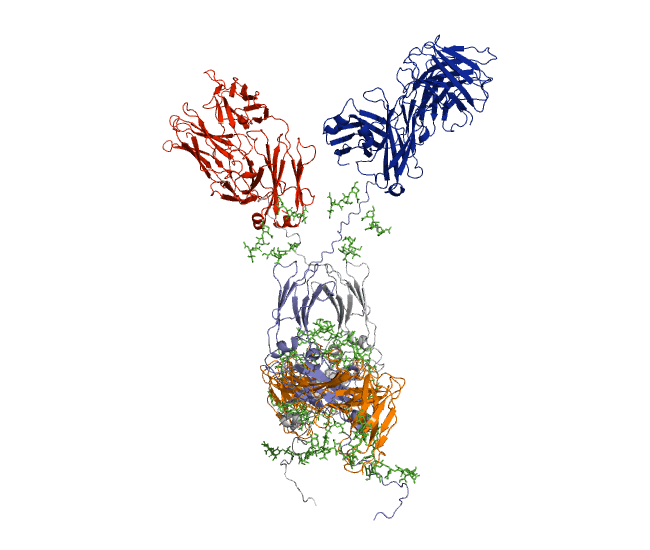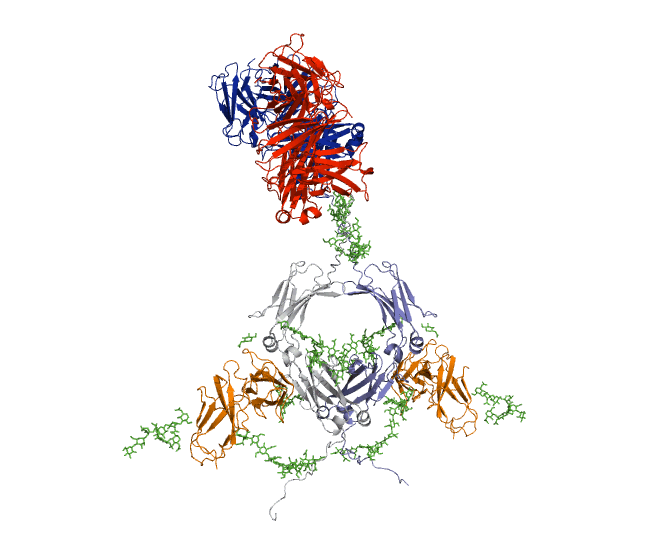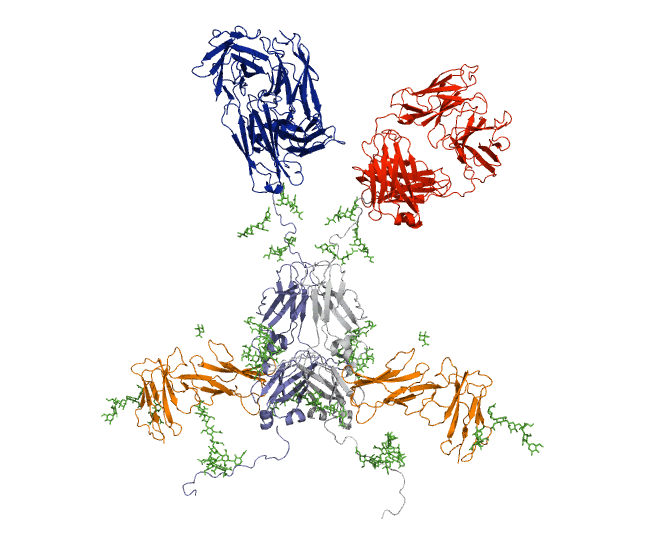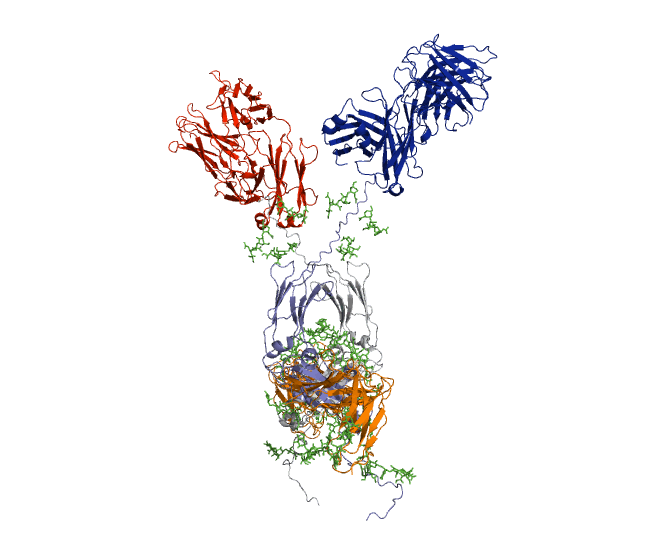
Our recent antibody results include the molecular dynamics modelling of human IgG1 antibody and its deglycosylation and likewise an IgG1-derived nanobody product C5-Fc based on SAXS and SANS data which provided the most detailed view of their solution structures to date (Spiteri et al., 2021; Gao et al., 2023). Similar analyses were performed for the human IgG2, IgG3 and IgG4 subclasses that accounted for their different immune reactivities (Hui et al., 2019, Spiteri et al, 2021). We have also looked at other antibody classes such as IgA (Hui et al., 2015) that is featured in the movie below.
Our most recent complement results from molecular dynamics modelling of SAXS data include the demonstration of significant flexibility in MBL, MASP, C4b and collagen that are relevant to lectin pathway activation (Fung et al., 2016; Nan et al., 2017, Walker et al., 2017, Iqbal et al., 2023).
Selected publications
Iqbal, H., Fung, K. W., Gor, J., Bishop, A. C., Makhatadze, G. I., Brodsky, B. and Perkins, S. J. (2023) A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin. J. Biol. Chem. 299, 102799. https://doi.org/10.1016/j.jbc.2022.102799 PubMed 36528062
Xu, Z., Spencer, H. J., Harris, V. A., and Perkins, S. J. (2023) An updated interactive database for 1692 genetic variants in coagulation Factor IX provides detailed insights into haemophilia B. J. Thromb. Haemost. 21, 1164-1176. https://doi.org/10.1016/j.jtha.2023.02.005 PubMed 36787808
Gao, X., Thrush, J. W., Gor, J., Naismith, J. H., Owens, R.J. & Perkins, S. J. (2023) The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions. J. Biol. Chem. 299, 105337. https://doi.org/10.1016/j.jbc.2023.105337 PubMed 37838175
