
Bioinformatics and Computational Biology
We are a computational biology laboratory with a particular interest in the genome-scale analysis of transcriptional regulation and evolution.
Our goal is to use genomic data to understand: (1) How is gene expression regulated; (2) How do these mechanisms control interesting biological behaviours; and (3) How does gene regulation affect evolution?
We use a genomic approach as it allows us to identify common principles that apply to most biological systems. Unique or unusual cases - which always occur in biology - could then be understood within this broader context.
Most of our work depends on publicly available data, but we also collaborate closely with a few experimental laboratories.
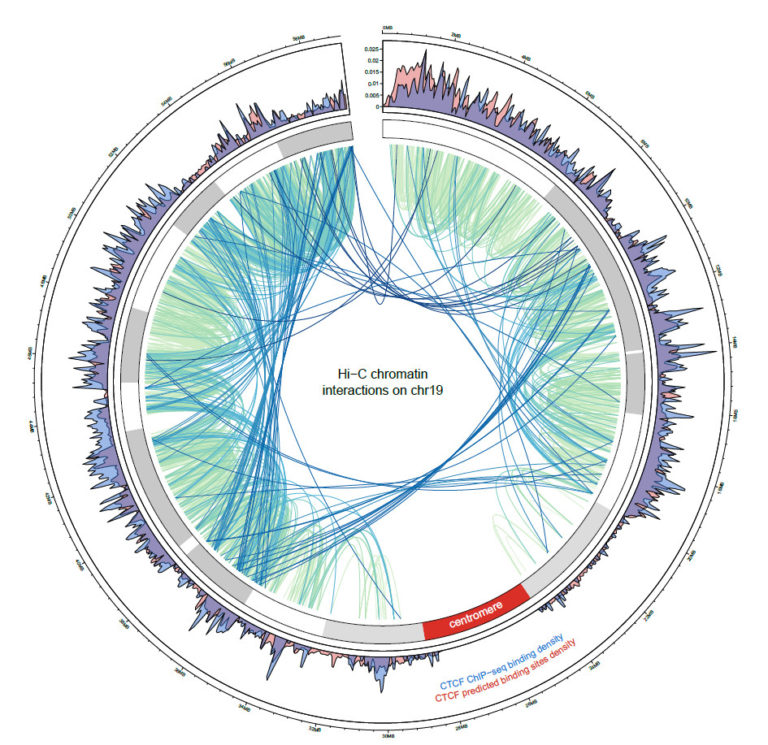
Selected publications
Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis
Poetsch, A.R., Boulton, S.J., Luscombe, N.M.
Poetsch, A.R., Boulton, S.J., Luscombe, N.M.
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
Mifsud, B., Tavares-Cadete, F., Young, A.N., Sugar, R., Schoenfelder, S., Ferreira, L., Wingett, S.W., Andrews, S., Grey, W., Ewels, P. A., Herman, B., Happe, S., Higgs, A., LeProust, E., Follows, G.A., Fraser, P., Luscombe, N.M., Osborne, C.S.
Nature Genetics (2015) 47 (6):598-606
Nature Genetics (2015) 47 (6):598-606
Evidence of non-random mutation rates suggests an evolutionary risk management strategy
Martincorena, I., Seshasayee, A.S., Luscombe, N.M.
Nature (2012) 485 (7396):95-98
Martincorena, I., Seshasayee, A.S., Luscombe, N.M.
Nature (2012) 485 (7396):95-98
