Computational Structural Biology
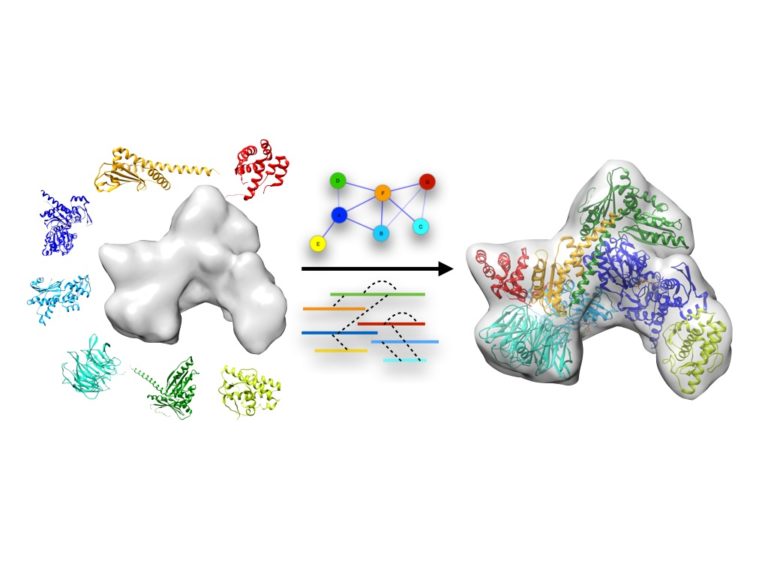
Our research is focused on the development of computational modelling approaches that integrate experimental data and bioinformatics to characterise the structure of macromolecular machines. Structural models are further analysed to understand their function and design new experiments. The group is particularly interested in applying these methods using data from cryo electron microscopy and mass spectrometry. In collaboration with other groups, specific systems we currently work on include herpesviruses, bacterial secretion systems, ligand-gated ion channels, and microtubule-associated proteins .
Selected publications
HVint: A strategy for identifying novel protein-protein interactions in herpes simplex virus type 1
Ashford, P., Hernandez, A., Greco, T.M., Buch, A., Sodeik, B., Cristea, I.M., Grünewald, K., Shepherd, A., Topf, M.
Molecular and Cellular Proteomics (2016) 15 (9):2939-2953
The importance of non-accessible crosslinks and solvent accessible surface distance in modelling proteins with restraints from crosslinking mass spectrometry
Bullock, J.M.A., Schwab, J., Thalassinos, K., Topf, M.
Molecular and Cellular Proteomics (2016) 15 (7):2491-2500
Refinement of atomic models in high resolution EM reconstructions using Flex-EM and local assessment.
Joseph, A.P., Malhotra, S., Burnley, T., Wood, C., Clare, D.K., Winn, M., Topf, M.
Methods (2016) 100:42-49
