Kristine Arnvig’s research group recently published a paper in the Nucleic Acids Journal. Full paper can be accessed here (doi.org/10.1093/nar/gkae338) .
Unexpected complexity of vitamin B12-sensing RNA elements in Mycobacterium tuberculosis.
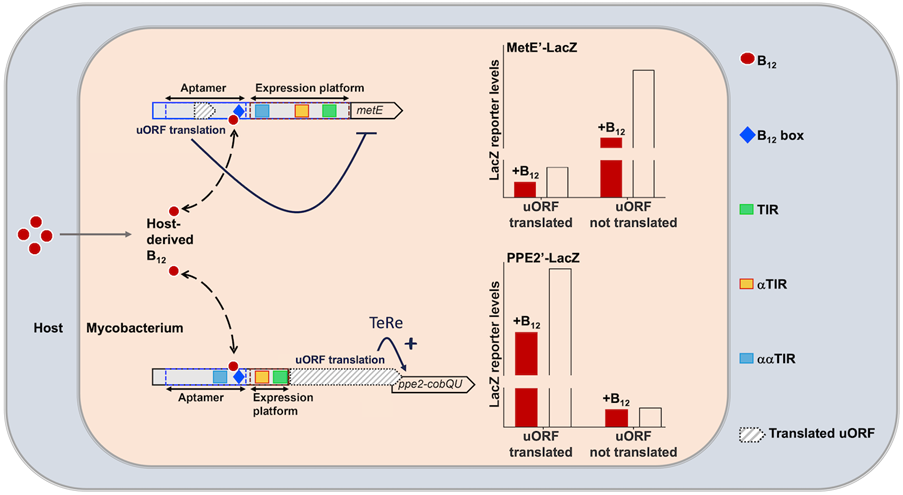
The bacterium Mycobacterium tuberculosis (Mtb) lacks the ability to produce vitamin B12, yet this co-factor plays a significant role in Mtb’s metabolism and gene regulation. Research conducted by Tine Arnvig’s team reveals an intriguing additional layer of control exerted by vitamin B12-sensing RNA elements, known as riboswitches, over metabolic and virulence genes in this pathogen. Beyond uncovering the B12-dependent inhibition of translation initiation, the team also uncovered two novel translated uORFs, which influence riboswitch regulation. In one instance, they identified a crucial translational link between the uORF and its downstream gene, facilitating translation re-initiation independently of Shine-Dalgarno sequences, alongside stop codon suppression, resulting in the synthesis of a frameshifted fusion protein. Essentially, Mtb demonstrates the capacity to generate similar proteins with differing N-termini, akin to eukaryotic alternative splicing. Lead author Dr. Terry Kipkorir remarks, “[This] represents yet another example of how Mtb breaks the mould in our understanding of gene expression control.’