
Cryo-electron microscopy of macromolecular machines
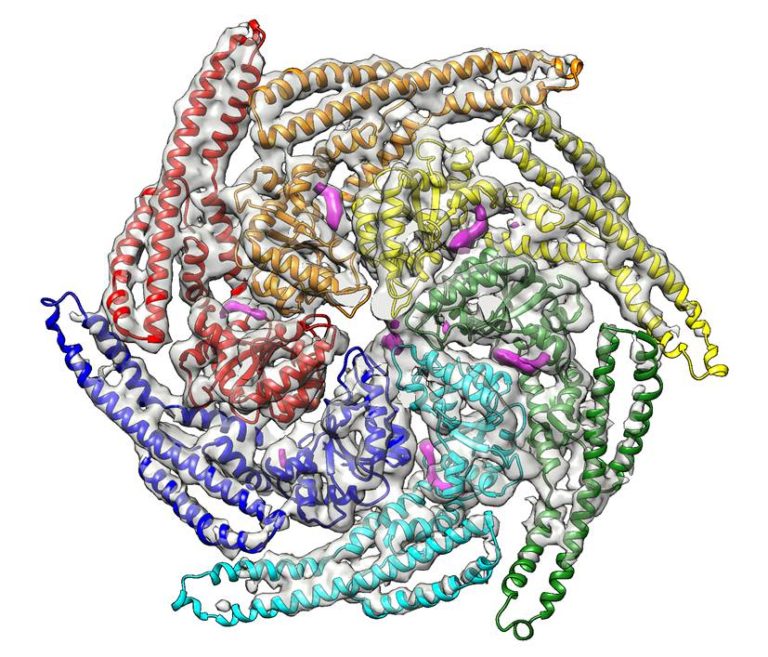
The Saibil group studies the operation of macromolecular machines using three-dimensional electron microscopy, using single particle cryo-electron microscopy and electron tomography of isolated complexes, irregular assemblies and cell sections. Our research projects are in the areas of molecular chaperones and assisted protein folding/unfolding, misfolding into amyloid, and protein refolding in membrane pore formation. In order to capture machine motions, we use statistical analysis to sort out different conformations in images of heterogeneous samples. On the cellular side, we are studying protein aggregation and disaggregation in situ. In addition, we study host cell interactions with intracellular pathogens, particularly the actions of malaria parasites on their host erythrocyte membranes and cytoskeleton. In collaborative work, we combine these EM studies with fluorescence and atomic force microscopies.
Selected publications
