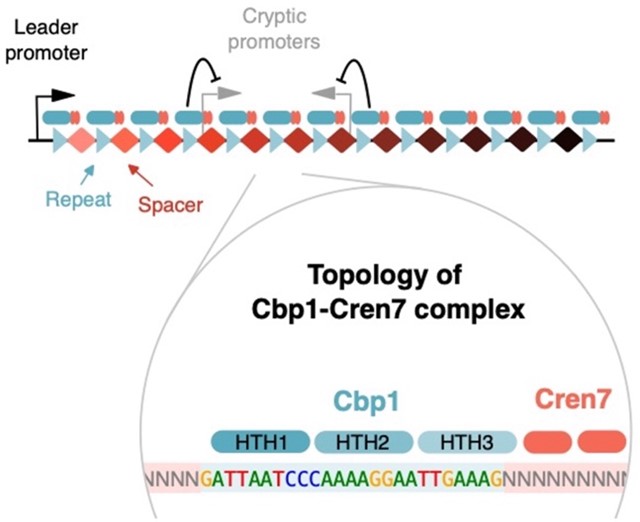
The prevalent state of DNA in all cells is chromatin that is made of protein-DNA complexes which are dynamic, complex and heterogenous. The exact composition of the chromatin determines its properties, how it regulates transcription and genome architecture. A breakthrough article by the RNAP laboratory at UCL published today in the journal Nature Communications describes how two chromatin proteins, Cbp1 and Cren7, collaborate to modulate gene regulation in opposing ways. While this type of specialised chromatin stimulates the expression of long crRNA arrays that facilitate adaptive immunity in archaea (‘CRISPR’), it cryptic promoters that frequently reside in the memory of the ‘immunity’ system, in the ‘CRISPR spacers’. This transcription interference by cryptic promoters limits how much spacer information can be stored in CRISPR arrays, or worse lead the malfunction of the system. Dr. Fabian Blombach from the ISMB RNAP laboratory, the lead author of the study, says ‘There is a lot of attention in the field to histones and their role in gene regulation, but we can learn so much from archaeal and bacterial chromatin proteins, including some of the ground rules that shape the interaction between chromatin proteins and transcription in all cellular life.’
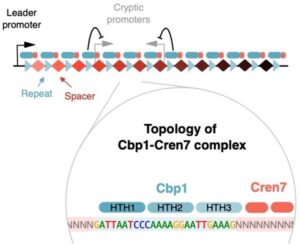
Sulfolobus Cbp1 and Cren7 form chimeric chromatin structures on large archaeal CRISPR arrays. Cbp1 confers sequence-specificity and interacts via its HTH3 domain interactions with Cren7. Together, they enhance leader promoters but repress CRISPR spacer-encoded cryptic promoters1. Archaea research in the RNAP lab is funded by the Wellcome Trust Investigator in Science Award (WT207446/Z/17/Z).
References
- Blombach, F., Sykora, M., Case, J., Feng, X., Baquero, D.P., Fouqueau, T., Phung, D.K., Barker, D., Krupovic, M., She, Q., and Werner, F. (2024). Cbp1 and Cren7 form chromatin-like structures that ensure efficient transcription of long CRISPR arrays. Nat Commun 15, 1620. 10.1038/s41467-024-45728-8.
Full paper can be read here.