
Professor and Chair of NMR Spectroscopy
Based at UCL
Combining Deep Learning and NMR Spectroscopy to characterise protein dynamics and interactions
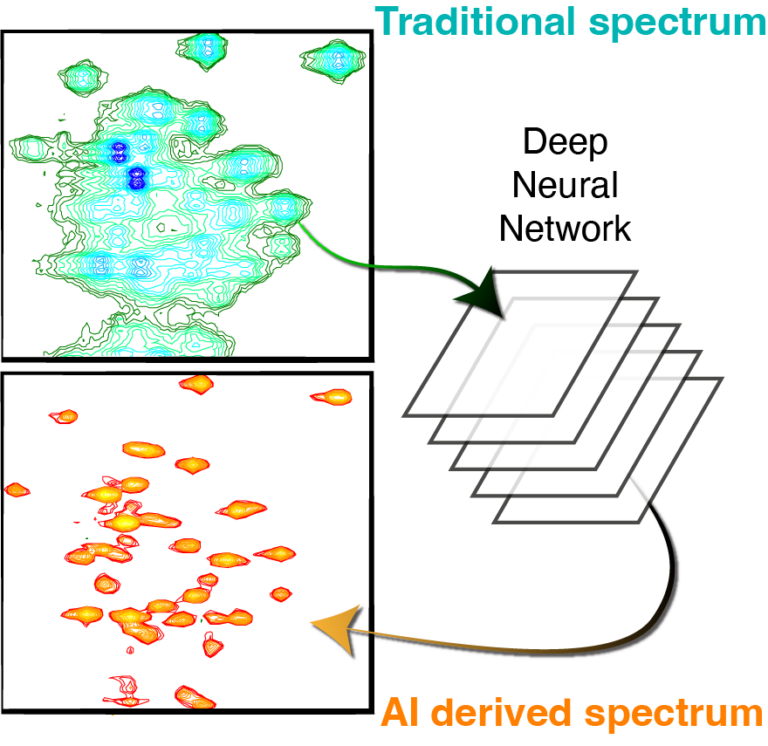
The Hansen Lab focuses on the study of protein dynamics using nuclear magnetic resonance (NMR) techniques. NMR method development is a major part of our research, including research into the analysis of NMR data using Deep Learning as well as the combination of NMR with computational tools. We are particularly interested in the dynamics, function, and regulations of human histone deacetylases (HDACs), domains of the von Willebrand Factor (vWF) and disordered viral proteins.
Selected publications
Intrinsic structural dynamics dictate enzymatic activity and inhibition
Vaibhav Kumar Shukla, Lucas Siemons, D Flemming Hansen
Proceedings of the National Academy of Sciences, 2023, https://doi.org/10.1073/pnas.2310910120
Virtual homonuclear decoupling in direct detection nuclear magnetic resonance experiments using deep neural networks
Gogulan Karunanithy, Harold W Mackenzie, D Flemming Hansen
Journal of the American Chemical Society, 2021, https://doi.org/10.1021/jacs.1c04010
FID-Net: A versatile deep neural network architecture for NMR spectral reconstruction and virtual decoupling
Gogulan Karunanithy, D Flemming Hansen
Journal of biomolecular NMR, 2021, https://doi.org/10.1007/s10858-021-00366-w
Characterising side chains in large proteins by protonless 13C-detected NMR spectroscopy
Ruth B Pritchard, D Flemming Hansen
Nature Communications, 2019, https://doi.org/10.1038/s41467-019-09743-4
